Identifying Optimal Proteins by Their Structure Using Graph Neural Networks
Arriola, M., Johnston, K., Frances, A.
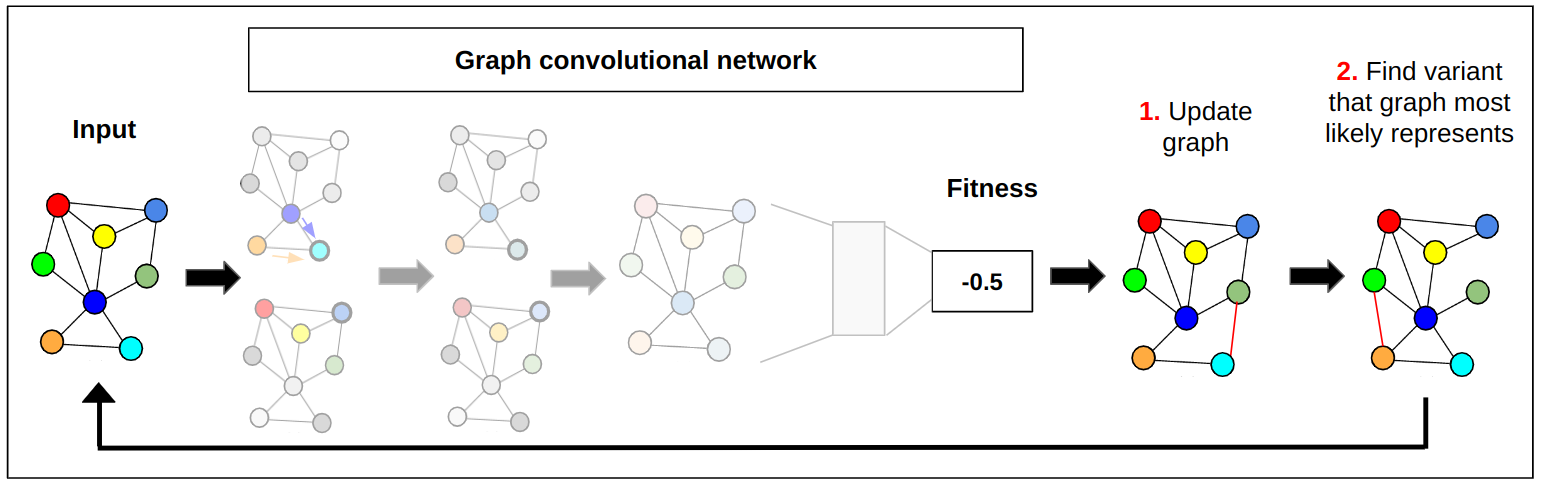
While standard protein representations depend on sequence, we hypothesize that protein structure provides richer information for the model to learn these properties. We represent each protein of interest as a graph, or a network of amino-acid connections in the protein, and implement a graph machine learning model to predict a protein’s fitness. We show that this structure-based model has superior performance to sequence-only approaches for fitness prediction. We further extend this model to automatically find the best protein for a given task by optimizing a protein’s graph representation.
